The FPU Recurrence Model of the Protein Synthesis
Andrey Berezin
Andrey Berezin*
EC- Leasing, DEMO-Center of IBM, Moscow, Russia
- *Corresponding Author:
- Andrey Berezin
EC-Leasing, DEMO-Center of IBM, Moscow, Russia.
Tel: 89671280467
E-mail: artparis@mail.ru
Received date: September 04, 2017; Accepted date: September 20, 2017; Published date: September 24, 2017
Citation: Berezin A (2017) The FPU Recurrence Model of the Protein Synthesis. Insights Med Phys. Vol. 2 No. 3:9
Abstract
The paper suggests a theoretical model of the physical mechanism of recognizing and joining of the transport RNA molecules with the information RNA molecules on the basis of the Fermi-Pasta-Ulam (FPU) recurrence and the group resonance phenomena. Both were experimentally observed in plasma dynamics. The suggested mathematical model represents two coupled nonlinear Shrodinger equations for the description of interaction between the FPU recurrence electric fields in the chains of the tRNA and iRNA molecules.
The results of numerical study of the model of dynamics of the tRNA and iRNA molecules in the intracellular solution allow making a conclusion that in a cell there exists a physical mechanism of recognizing, attracting and repelling between the tRNA and iRNA molecules, providing the synthesis of protein. This mechanism is based on the FPU recurrence, whose spectrum structure gives a pattern – matrix for building a protein. Such resonant dynamics is generally characteristic for the dynamics of interaction between the FPU recurrences, in particular the elementary FPU recurrence of the tRNA molecule electrical field and full FPU recurrence of the iRNA molecule electrical field.
Moreover, the suggested physical mechanism allows offering a method of external influence on a cell aiming at acceleration of the protein synthesis in it by the applying electromagnetic fields in a form of the FPU recurrence spectrum.
Keywords
Dynamics; tRNA; FPU model; Acceleration; Spectrum
Introduction
As it is known, the genetic information is transferred to ribosomes via molecules of the information RNA. The latter is synthesized on a gene along one of two spirals of the DNA molecule on the basis of the complementarities principle of the bases. One information RNA molecule contains the sequence of nucleotides, complementary to one that served as a matrix, and is identical to the sequence of the second spiral of the DNA molecule.
Information RNA is attached to ribosomes. Various amino acids with the help of specific enzymes join the transport RNA molecules, specific to each amino acid. The transport RNA with the amino acid attached to it "recognizes" among the sequence of the information RNA nucleotides the triplet encoding the amino acid being carried. The "recognizing" of it is carried out thanks to the complementary triplet –anticodon, allowing them to join temporarily. If the codon is complementary to the information RNA triplet, the amino acid is disconnected from the transport ÃÆÃÂÃâàÃÆÃÂÃâÃÂÃÆÃÂÃâà ¡ and joins the growing protein sequence. However, the described sophisticated mechanism, as well as many other mechanisms of biological processes, which have only verbal descriptions of the sequences of actions are not yet physically explained. The physical bases of the processes which don’t contain any conscious actions, in particular, "recognizing" and joining the transport RNA, having the amino acid, with the molecule of the information RNA among chaotically moving molecules of the intracellular solution is not found out yet.
Model of Protein Sythesis
The purpose of the paper was to develop a theoretical model of the physical mechanism of recognizing and joining of the transport RNA molecules with the information RNA molecules on the basis of the FPU recurrence [1] and the group resonance phenomena [2]. The group resonance approach has been used in the theory of plasma for describing the dynamics of the particle movement in the field of a wave packet. This problem comprises many characteristic features typical for various physical situations [2]. The formal description of the problem is expressed as follows:
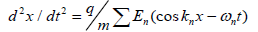 (1)
(1)
Where x-is the coordinate, q-is the particle charge, m-is the particle mass, Σ -is the wave packet of the field acting on the particle.
Together with this, the FPU recurrence [1], proved to be the main characteristic property of distributed dynamic systems including plasma, also looks as a useful tool for modeling the process under consideration.
As it was shown earlier an elementary FPU recurrence develops in a chain consisting of three coupled masses [3]. Representing the tRNA molecule as an elementary FPU recurrence chain and the iRNA molecule as the classical FPU recurrence chain [1], we can take an advantage of the approach suggested by Berman and Kolovsky [4] who have shown, that the dynamics of the FPU chains can be described within the framework of Nonlinear Shrodinger Equation (NSE). Besides the RNA electrical field dynamics can be given within the frame of coupled sine- Gordon and NSE equations forming the FPU recurrence in the cell of neuron [5]. Using the equation (1) for the description of the group resonance, and also the form of the Shrodinger equation with the dispersing potential, suggested by Bishop [6], we can write down the process of interaction between the tRNA and iRNA molecules electric fields in the form of the FPU recurrences raised in the quasi periodical structures of these molecules by the energy ÃÆÃÂÃâà ¡ÃÆÃÂÃââ of the intracellular liquid, as the perturbed nonlinear Shrodinger equations. The hypothetical mechanism of extracting energy of random fluctuations of the medium and transforming it into the energy of oscillations in the FPU chains placed into this medium, for the first time was suggested by Frolich [7] and then was confirmed mathematically as a result of numerical study of the elementary model of the FPU recurrence [8] and then was proved experimentally through developing the physical model of that process [9].
Accounting that the tRNA molecule represents a closed FPU chain, and the iRNA molecule represents the FPU chain with open ends, he NSE equations for the description of interaction of the electric FPU fields in these chains will contain the packets having odd spatial harmonics (iRNA) [10] and even spatial harmonics (tRNA) [11] as perturbing functions:
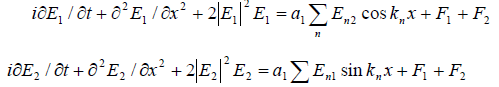 (2)
(2)
where a1 and a2 are the intensities of the tRNA and iRNA electric fields correspondingly, a1 and a2 are constants, Σ E2n and Σ E2n are the wave packets of the electric fields mutually acting on the iRNA and tRNA molecules correspondingly.
F1 and F2 are random functions describing correspondingly the high- and low-frequency chaotic fluctuations in the intracellular liquid.
Following Bishop's work [4], the solutions of the system (2) can be given as follows:
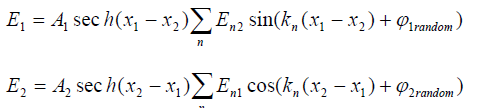 (3)
(3)
where 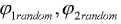 are the random phases
are the random phases
Using a reduced version of (1), the equations of a group resonance have been replaced by the equations of nonlinear pendulum with a changing frequency [1] in which the solutions of the system (3) have been included into the conservative members of the reduced equations. Then having a purpose of simplification of the obtained system, the perturbing wave packets have been left with only two harmonics – the odd ones - for a packet of the iRNA and the even ones - for a packet of the tRNA. The system looks as follows:
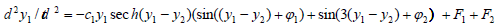 (4)
(4)
where y1,y2 are the coordinates of the molecules tRNA and iRNA correspondingly, ÃÆÃÂÃâââ¬Â¢1 ,ÃÆÃÂÃâââ¬Â¢2 ,ÃÆÃÂÃâââ¬Â¢3 ,ÃÆÃÂÃâââ¬Â¢4 are the constants, ÃÆÃÂÃâââ¬Â¢1 ,ÃÆÃÂÃâââ¬Â¢2 ,ÃÆÃÂÃâââ¬Â¢3 ,ÃÆÃÂÃâââ¬Â¢4 are random functions describing correspondingly the high- and lowfrequency chaotic fluctuations in the intracellular liquid.
Results and Discussion
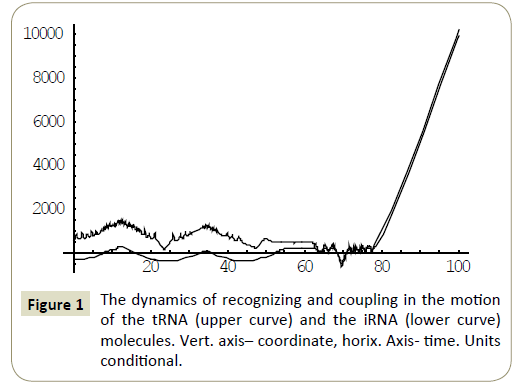
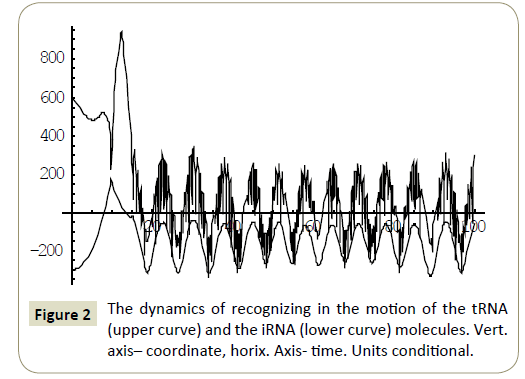
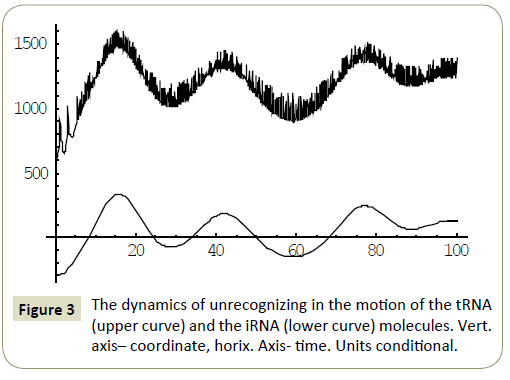
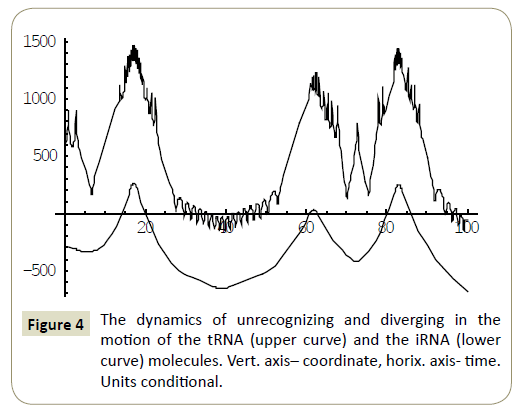
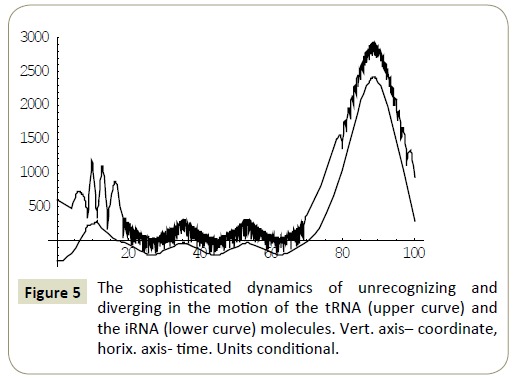
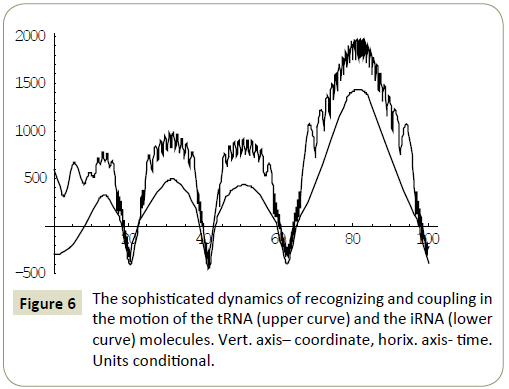
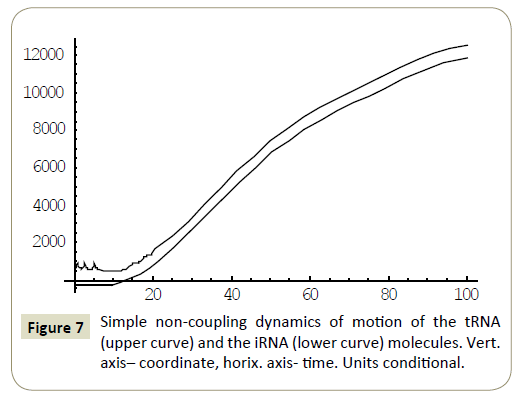
Numerical study of the system (4) has shown, that within the framework of the proposed model, the RNA molecules proved to be able to “recognize” each other and then couple or divert from each other as a result of “unrecognition” depending on the certain phase combinations ( ÃÆÃÂÃâââ¬Â¢1 ,ÃÆÃÂÃâââ¬Â¢2 ,ÃÆÃÂÃâââ¬Â¢3 ,ÃÆÃÂÃâââ¬Â¢4 ) of the wave packets harmonics of the tRNA and iRNA molecules. Figures 1 and 2 illustrate the process of recognition and coupling of the RNA molecules. Figures 3 and 4 demonstrate divergence of the RNA molecules after “unrecognition”. The processes of recognition, attraction and repelling proved to be sophisticated ones. Figures 5 and 6 show some of them. Figure 7 shows a simple dynamics of the divergence process between the RNA molecules.
Conclusion
The results of numerical study of the model of dynamics of the tRNA and iRNA molecules in the intracellular solution allow making a conclusion that in a cell there exists a physical mechanism of recognizing, attracting and repelling between the tRNA and iRNA molecules, providing the synthesis of protein. Such resonant dynamics is characteristic for the dynamics of interaction between the FPU recurrences [12]: the elementary FPU recurrence of the tRNA molecule electrical field and the full FPU recurrence of the iRNA molecule electrical field.
Moreover, the suggested physical mechanism allows offering a method of external influence on a cell aiming at acceleration of the protein synthesis in it by the electromagnetic fields in a form of the FPU recurrence spectrum. The problem of breakage of the proposed mechanism was not considered but a probable fatal mode can occur in the wrongly differentiated cell in a form of a self parametric excitation of the protein assembling process when a distorted protein molecule will be constantly reproduced on a tumor iRNA at a much higher speed than that of a normal protein molecule triggering the process of self-sustained tumor cells reduplication through production of wrong proteins.
References
- Fermi E, Pasta J, Ulam S (1995) In: Collected Papers of E. Fermi. 2: 978.
- Zaslavsky GM, Chernikov (1985) AA Group resonance, universal mapping and stochastic dynamics of particles. Sov Phys JETP 89: 1632-1647.
- Berezin AA (1989) Can an electromagnetic field exist in a form of Fermi-Pasta-Ulam recurrence? J Phys D Appl Phys 22: 577-589.
- Berman GP, Kolovskii AR (1984) The limit of stochasticity for a one-dimensional chain of interacting oscillators. Sov Phys JETP 60: 1116.
- Berezin AA (1999) Neural RNA as a principal dynamic information carrier in a neuron. SPIE 3812: 1325-1337.
- Bishop AR (1978) Solitons and physical perturbations. In Solitons in Action. Ed. Academic Press. NY, USA. pp. 72-102.
- Frolich H (1968) Storage of light energy and photosynthesis. Phys Lett 26:402.
- Berezin AA, Pavel BK, Erast UA (1988) Theoretical simulation of a coherent coupled electromagnetic wave structure induced by the white noise. Physica Scripta 38: 719-720.
- Berezin AA, Pavel BK, Erast UA (1989) Experimental investigation of a coherent coupled electromagnetic wave excited in the plasma of a bulb with a tungsten filament. Physica Scripta 39: 474-482.
- Yuen HC, Lake BM (1978) Nonlinear wave concept applied to deep water waves. in Solitons in Action. Academic Press, NY, USA. pp. 103-137.
- Zubusky NJ, Kruskal MD (1965) Interaction of solitons in a collision less plasma and the recurrence of initial Statesphys. Rev Lett 15:240.
- Berezin AA (2004) Resonant interaction between the Fermi-Pasta-Ulam recurrences. Bul Lebedev Phys Ins. (Russian Academy of Sciences). Allerton Press NY, USA. pp. 4: 41-50.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences